utilities¶
This module contains various utility methods that support functionality in other modules. The multiprocessing functionality also exists in this module, though that might change with time.
binner(signal, times, bins) |
|
brute_force_search(grids, bounds, Ns, data, ...) |
A generic brute-force grid-search error minimization function. |
cartes_to_polar(cartes) |
Assumes that the 0th and 1st parameters are cartesian x and y |
double_gamma_hrf(delay, tr[, fptr, integrator]) |
The double gamma hemodynamic reponse function (HRF). |
error_function(parameters, bounds, data, ...) |
A generic error function with bounding. |
find_files(directory, pattern) |
|
gaussian_2D(X, Y, x0, y0, sigma_x, sigma_y, ...) |
|
generate_shared_array(unshared_arr, dtype) |
Creates synchronized shared arrays from numpy arrays. |
gradient_descent_search(parameters, bounds, ...) |
A generic gradient-descent error minimization function. |
make_nifti(data[, grid_parent]) |
|
multiprocess_bundle(Fit, model, data, grids, ...) |
|
normalize(array[, imin, imax, axis]) |
A short-hand function for normalizing an array to a desired range. |
parallel_fit(args) |
This is a convenience function for parallelizing the fitting procedure. |
peakdet(v, delta[, x]) |
Converted from MATLAB script at http://billauer.co.il/peakdet.html |
percent_change(ts[, ax]) |
Returns the % signal change of each point of the times series |
randn(d0, d1, ..., dn) |
Return a sample (or samples) from the “standard normal” distribution. |
recast_estimation_results(output, grid_parent) |
|
seed([seed]) |
Seed the generator. |
spm_hrf(delay, TR) |
An implementation of spm_hrf.m from the SPM distribution |
zscore(time_series[, axis]) |
Returns the z-score of each point of the time series along a given axis of the array time_series. |
Array¶
-
popeye.utilities.Array(typecode_or_type, size_or_initializer, **kwds)¶ Returns a synchronized shared array
arange¶
-
popeye.utilities.arange([start, ]stop, [step, ]dtype=None)¶ Return evenly spaced values within a given interval.
Values are generated within the half-open interval
[start, stop)(in other words, the interval including start but excluding stop). For integer arguments the function is equivalent to the Python built-in range function, but returns an ndarray rather than a list.When using a non-integer step, such as 0.1, the results will often not be consistent. It is better to use
linspacefor these cases.- start : number, optional
- Start of interval. The interval includes this value. The default start value is 0.
- stop : number
- End of interval. The interval does not include this value, except in some cases where step is not an integer and floating point round-off affects the length of out.
- step : number, optional
- Spacing between values. For any output out, this is the distance
between two adjacent values,
out[i+1] - out[i]. The default step size is 1. If step is specified, start must also be given. - dtype : dtype
- The type of the output array. If dtype is not given, infer the data type from the other input arguments.
- arange : ndarray
Array of evenly spaced values.
For floating point arguments, the length of the result is
ceil((stop - start)/step). Because of floating point overflow, this rule may result in the last element of out being greater than stop.
linspace : Evenly spaced numbers with careful handling of endpoints. ogrid: Arrays of evenly spaced numbers in N-dimensions. mgrid: Grid-shaped arrays of evenly spaced numbers in N-dimensions.
>>> np.arange(3) array([0, 1, 2]) >>> np.arange(3.0) array([ 0., 1., 2.]) >>> np.arange(3,7) array([3, 4, 5, 6]) >>> np.arange(3,7,2) array([3, 5])
array¶
-
popeye.utilities.array(object, dtype=None, copy=True, order=None, subok=False, ndmin=0)¶ Create an array.
- object : array_like
- An array, any object exposing the array interface, an object whose __array__ method returns an array, or any (nested) sequence.
- dtype : data-type, optional
- The desired data-type for the array. If not given, then the type will be determined as the minimum type required to hold the objects in the sequence. This argument can only be used to ‘upcast’ the array. For downcasting, use the .astype(t) method.
- copy : bool, optional
- If true (default), then the object is copied. Otherwise, a copy will only be made if __array__ returns a copy, if obj is a nested sequence, or if a copy is needed to satisfy any of the other requirements (dtype, order, etc.).
- order : {‘C’, ‘F’, ‘A’}, optional
- Specify the order of the array. If order is ‘C’, then the array will be in C-contiguous order (last-index varies the fastest). If order is ‘F’, then the returned array will be in Fortran-contiguous order (first-index varies the fastest). If order is ‘A’ (default), then the returned array may be in any order (either C-, Fortran-contiguous, or even discontiguous), unless a copy is required, in which case it will be C-contiguous.
- subok : bool, optional
- If True, then sub-classes will be passed-through, otherwise the returned array will be forced to be a base-class array (default).
- ndmin : int, optional
- Specifies the minimum number of dimensions that the resulting array should have. Ones will be pre-pended to the shape as needed to meet this requirement.
- out : ndarray
- An array object satisfying the specified requirements.
empty, empty_like, zeros, zeros_like, ones, ones_like, fill
>>> np.array([1, 2, 3]) array([1, 2, 3])
Upcasting:
>>> np.array([1, 2, 3.0]) array([ 1., 2., 3.])
More than one dimension:
>>> np.array([[1, 2], [3, 4]]) array([[1, 2], [3, 4]])
Minimum dimensions 2:
>>> np.array([1, 2, 3], ndmin=2) array([[1, 2, 3]])
Type provided:
>>> np.array([1, 2, 3], dtype=complex) array([ 1.+0.j, 2.+0.j, 3.+0.j])
Data-type consisting of more than one element:
>>> x = np.array([(1,2),(3,4)],dtype=[('a','<i4'),('b','<i4')]) >>> x['a'] array([1, 3])
Creating an array from sub-classes:
>>> np.array(np.mat('1 2; 3 4')) array([[1, 2], [3, 4]])
>>> np.array(np.mat('1 2; 3 4'), subok=True) matrix([[1, 2], [3, 4]])
asarray¶
-
popeye.utilities.asarray(a, dtype=None, order=None)¶ Convert the input to an array.
- a : array_like
- Input data, in any form that can be converted to an array. This includes lists, lists of tuples, tuples, tuples of tuples, tuples of lists and ndarrays.
- dtype : data-type, optional
- By default, the data-type is inferred from the input data.
- order : {‘C’, ‘F’}, optional
- Whether to use row-major (C-style) or column-major (Fortran-style) memory representation. Defaults to ‘C’.
- out : ndarray
- Array interpretation of a. No copy is performed if the input is already an ndarray. If a is a subclass of ndarray, a base class ndarray is returned.
asanyarray : Similar function which passes through subclasses. ascontiguousarray : Convert input to a contiguous array. asfarray : Convert input to a floating point ndarray. asfortranarray : Convert input to an ndarray with column-major
memory order.asarray_chkfinite : Similar function which checks input for NaNs and Infs. fromiter : Create an array from an iterator. fromfunction : Construct an array by executing a function on grid
positions.Convert a list into an array:
>>> a = [1, 2] >>> np.asarray(a) array([1, 2])
Existing arrays are not copied:
>>> a = np.array([1, 2]) >>> np.asarray(a) is a True
If dtype is set, array is copied only if dtype does not match:
>>> a = np.array([1, 2], dtype=np.float32) >>> np.asarray(a, dtype=np.float32) is a True >>> np.asarray(a, dtype=np.float64) is a False
Contrary to asanyarray, ndarray subclasses are not passed through:
>>> issubclass(np.matrix, np.ndarray) True >>> a = np.matrix([[1, 2]]) >>> np.asarray(a) is a False >>> np.asanyarray(a) is a True
brute¶
-
popeye.utilities.brute(func, ranges, args=(), Ns=20, full_output=0, finish=<function fmin>, disp=False)¶ Minimize a function over a given range by brute force.
- func : callable
f(x,*args) - Objective function to be minimized.
- ranges : tuple
- Each element is a tuple of parameters or a slice object to
be handed to
numpy.mgrid. - args : tuple
- Extra arguments passed to function.
- Ns : int
- Default number of samples, if those are not provided.
- full_output : bool
- If True, return the evaluation grid.
- finish : callable, optional
- An optimization function that is called with the result of brute force minimization as initial guess. finish should take the initial guess as positional argument, and take take args, full_output and disp as keyword arguments. See Notes for more details.
- disp : bool, optional
- Set to True to print convergence messages.
- x0 : ndarray
- Value of arguments to func, giving minimum over the grid.
- fval : int
- Function value at minimum.
- grid : tuple
- Representation of the evaluation grid. It has the same length as x0.
- Jout : ndarray
- Function values over grid:
Jout = func(*grid).
The range is respected by the brute force minimization, but if the finish keyword specifies another optimization function (including the default fmin), the returned value may still be (just) outside the range. In order to ensure the range is specified, use
finish=None.- func : callable
brute_force_search¶
-
popeye.utilities.brute_force_search(grids, bounds, Ns, data, error_function, objective_function, verbose)¶ A generic brute-force grid-search error minimization function.
The user specifies an objective_function and the corresponding args for generating a model prediction. The brute_force_search uses search_bounds to dictate the bounds for evenly sample the parameter space.
In addition, the user must also supply fit_bounds, containing pairs of upper and lower bounds for each of the values in parameters. If fit_bounds is specified, the error minimization procedure in error_function will return an Inf whether the parameters exceed the minimum or maxmimum values specified in fit_bounds.
The output of brute_force_search can be used as a seed-point for the fine-tuned solutions from gradient_descent_search.
- args : tuple
- Arguments to objective_function that yield a model prediction.
- grids : tuple
A tuple indicating the search space for the brute-force grid-search. The tuple contains pairs of upper and lower bounds for exploring a given dimension. For example grids=((-10,10),(0,5),) will search the first dimension from -10 to 10 and the second from 0 to 5. These values cannot be None.
For more information, see scipy.optimize.brute.
- bounds : tuple
- A tuple containing the upper and lower bounds for each parameter in parameters. If a parameter is not bounded, simply use None. For example, fit_bounds=((0,None),(-10,10),) would bound the first parameter to be any positive number while the second parameter would be bounded between -10 and 10.
- Ns : int
Number of samples per stimulus dimension to sample during the ballpark search.
For more information, see scipy.optimize.brute.
- data : ndarray
- The actual, measured time-series against which the model is fit.
- error_function : callable
- The error function that relates the model prediction to the measure data. The error function returns a float that represents the residual sum of squared errors between the prediction and the data.
- objective_function : callable
- The objective function that takes parameters and args and proceduces a model time-series.
- estimate : tuple
- The model solution given parameters and objective_function.
cartes_to_polar¶
-
popeye.utilities.cartes_to_polar(cartes)¶ Assumes that the 0th and 1st parameters are cartesian x and y
det¶
-
popeye.utilities.det(a, overwrite_a=False, check_finite=True)¶ Compute the determinant of a matrix
The determinant of a square matrix is a value derived arithmetically from the coefficients of the matrix.
The determinant for a 3x3 matrix, for example, is computed as follows:
a b c d e f = A g h i det(A) = a*e*i + b*f*g + c*d*h - c*e*g - b*d*i - a*f*h
- a : (M, M) array_like
- A square matrix.
- overwrite_a : bool
- Allow overwriting data in a (may enhance performance).
- check_finite : boolean, optional
- Whether to check the input matrixes contain only finite numbers. Disabling may give a performance gain, but may result to problems (crashes, non-termination) if the inputs do contain infinities or NaNs.
- det : float or complex
- Determinant of a.
The determinant is computed via LU factorization, LAPACK routine z/dgetrf.
>>> a = np.array([[1,2,3],[4,5,6],[7,8,9]]) >>> linalg.det(a) 0.0 >>> a = np.array([[0,2,3],[4,5,6],[7,8,9]]) >>> linalg.det(a) 3.0
diagonal¶
-
popeye.utilities.diagonal(a, offset=0, axis1=0, axis2=1)¶ Return specified diagonals.
If a is 2-D, returns the diagonal of a with the given offset, i.e., the collection of elements of the form
a[i, i+offset]. If a has more than two dimensions, then the axes specified by axis1 and axis2 are used to determine the 2-D sub-array whose diagonal is returned. The shape of the resulting array can be determined by removing axis1 and axis2 and appending an index to the right equal to the size of the resulting diagonals.In versions of NumPy prior to 1.7, this function always returned a new, independent array containing a copy of the values in the diagonal.
In NumPy 1.7 and 1.8, it continues to return a copy of the diagonal, but depending on this fact is deprecated. Writing to the resulting array continues to work as it used to, but a FutureWarning is issued.
Starting in NumPy 1.9 it returns a read-only view on the original array. Attempting to write to the resulting array will produce an error.
In some future release, it will return a read/write view and writing to the returned array will alter your original array. The returned array will have the same type as the input array.
If you don’t write to the array returned by this function, then you can just ignore all of the above.
If you depend on the current behavior, then we suggest copying the returned array explicitly, i.e., use
np.diagonal(a).copy()instead of justnp.diagonal(a). This will work with both past and future versions of NumPy.- a : array_like
- Array from which the diagonals are taken.
- offset : int, optional
- Offset of the diagonal from the main diagonal. Can be positive or negative. Defaults to main diagonal (0).
- axis1 : int, optional
- Axis to be used as the first axis of the 2-D sub-arrays from which the diagonals should be taken. Defaults to first axis (0).
- axis2 : int, optional
- Axis to be used as the second axis of the 2-D sub-arrays from which the diagonals should be taken. Defaults to second axis (1).
- array_of_diagonals : ndarray
- If a is 2-D and not a matrix, a 1-D array of the same type as a containing the diagonal is returned. If a is a matrix, a 1-D array containing the diagonal is returned in order to maintain backward compatibility. If the dimension of a is greater than two, then an array of diagonals is returned, “packed” from left-most dimension to right-most (e.g., if a is 3-D, then the diagonals are “packed” along rows).
- ValueError
- If the dimension of a is less than 2.
diag : MATLAB work-a-like for 1-D and 2-D arrays. diagflat : Create diagonal arrays. trace : Sum along diagonals.
>>> a = np.arange(4).reshape(2,2) >>> a array([[0, 1], [2, 3]]) >>> a.diagonal() array([0, 3]) >>> a.diagonal(1) array([1])
A 3-D example:
>>> a = np.arange(8).reshape(2,2,2); a array([[[0, 1], [2, 3]], [[4, 5], [6, 7]]]) >>> a.diagonal(0, # Main diagonals of two arrays created by skipping ... 0, # across the outer(left)-most axis last and ... 1) # the "middle" (row) axis first. array([[0, 6], [1, 7]])
The sub-arrays whose main diagonals we just obtained; note that each corresponds to fixing the right-most (column) axis, and that the diagonals are “packed” in rows.
>>> a[:,:,0] # main diagonal is [0 6] array([[0, 2], [4, 6]]) >>> a[:,:,1] # main diagonal is [1 7] array([[1, 3], [5, 7]])
diff¶
-
popeye.utilities.diff(a, n=1, axis=-1)¶ Calculate the n-th discrete difference along given axis.
The first difference is given by
out[n] = a[n+1] - a[n]along the given axis, higher differences are calculated by using diff recursively.- a : array_like
- Input array
- n : int, optional
- The number of times values are differenced.
- axis : int, optional
- The axis along which the difference is taken, default is the last axis.
- diff : ndarray
- The n-th differences. The shape of the output is the same as a except along axis where the dimension is smaller by n.
.
gradient, ediff1d, cumsum
>>> x = np.array([1, 2, 4, 7, 0]) >>> np.diff(x) array([ 1, 2, 3, -7]) >>> np.diff(x, n=2) array([ 1, 1, -10])
>>> x = np.array([[1, 3, 6, 10], [0, 5, 6, 8]]) >>> np.diff(x) array([[2, 3, 4], [5, 1, 2]]) >>> np.diff(x, axis=0) array([[-1, 2, 0, -2]])
dot¶
-
popeye.utilities.dot(a, b, out=None)¶ Dot product of two arrays.
For 2-D arrays it is equivalent to matrix multiplication, and for 1-D arrays to inner product of vectors (without complex conjugation). For N dimensions it is a sum product over the last axis of a and the second-to-last of b:
dot(a, b)[i,j,k,m] = sum(a[i,j,:] * b[k,:,m])
- a : array_like
- First argument.
- b : array_like
- Second argument.
- out : ndarray, optional
- Output argument. This must have the exact kind that would be returned if it was not used. In particular, it must have the right type, must be C-contiguous, and its dtype must be the dtype that would be returned for dot(a,b). This is a performance feature. Therefore, if these conditions are not met, an exception is raised, instead of attempting to be flexible.
- output : ndarray
- Returns the dot product of a and b. If a and b are both scalars or both 1-D arrays then a scalar is returned; otherwise an array is returned. If out is given, then it is returned.
- ValueError
- If the last dimension of a is not the same size as the second-to-last dimension of b.
vdot : Complex-conjugating dot product. tensordot : Sum products over arbitrary axes. einsum : Einstein summation convention. matmul : ‘@’ operator as method with out parameter.
>>> np.dot(3, 4) 12
Neither argument is complex-conjugated:
>>> np.dot([2j, 3j], [2j, 3j]) (-13+0j)
For 2-D arrays it is the matrix product:
>>> a = [[1, 0], [0, 1]] >>> b = [[4, 1], [2, 2]] >>> np.dot(a, b) array([[4, 1], [2, 2]])
>>> a = np.arange(3*4*5*6).reshape((3,4,5,6)) >>> b = np.arange(3*4*5*6)[::-1].reshape((5,4,6,3)) >>> np.dot(a, b)[2,3,2,1,2,2] 499128 >>> sum(a[2,3,2,:] * b[1,2,:,2]) 499128
double_gamma_hrf¶
-
popeye.utilities.double_gamma_hrf(delay, tr, fptr=1.0, integrator=<function trapz>)¶ The double gamma hemodynamic reponse function (HRF). The user specifies only the delay of the peak and undershoot. The delay shifts the peak and undershoot by a variable number of seconds. The other parameters are hardcoded. The HRF delay is modeled for each voxel independently. The form of the HRF and the hardcoded values are based on previous work [1]_.
- delay : float
- The delay of the HRF peak and undershoot.
- tr : float
- The length of the repetition time in seconds.
- fptr : float
- The number of stimulus frames per reptition time. For a 60 Hz projector and with a 1 s repetition time, the fptr would be equal to 60. It is possible that you will bin all the frames in a single TR, in which case fptr equals 1.
- integrator : callable
- The integration function for normalizing the units of the HRF so that the area under the curve is the same for differently delayed HRFs. Set integrator to None to turn off normalization.
- hrf : ndarray
- The hemodynamic response function to convolve with the stimulus timeseries.
[1] Glover, GH (1999) Deconvolution of impulse response in event related BOLD fMRI. NeuroImage 9, 416-429.
error_function¶
-
popeye.utilities.error_function(parameters, bounds, data, objective_function, verbose)¶ A generic error function with bounding.
- parameters : tuple
- A tuple of values representing a model setting.
- args : tuple
- Extra arguments to objective_function beyond those in parameters.
- data : ndarray
- The actual, measured time-series against which the model is fit.
- objective_function : callable
- The objective function that takes parameters and args and proceduces a model time-series.
- debug : bool
- Useful for debugging a model, will print the parameters and error.
- error : float
- The residual sum of squared errors between the prediction and data.
fmin¶
-
popeye.utilities.fmin(func, x0, args=(), xtol=0.0001, ftol=0.0001, maxiter=None, maxfun=None, full_output=0, disp=1, retall=0, callback=None)¶ Minimize a function using the downhill simplex algorithm.
This algorithm only uses function values, not derivatives or second derivatives.
- func : callable func(x,*args)
- The objective function to be minimized.
- x0 : ndarray
- Initial guess.
- args : tuple, optional
- Extra arguments passed to func, i.e.
f(x,*args). - callback : callable, optional
- Called after each iteration, as callback(xk), where xk is the current parameter vector.
- xtol : float, optional
- Relative error in xopt acceptable for convergence.
- ftol : number, optional
- Relative error in func(xopt) acceptable for convergence.
- maxiter : int, optional
- Maximum number of iterations to perform.
- maxfun : number, optional
- Maximum number of function evaluations to make.
- full_output : bool, optional
- Set to True if fopt and warnflag outputs are desired.
- disp : bool, optional
- Set to True to print convergence messages.
- retall : bool, optional
- Set to True to return list of solutions at each iteration.
- xopt : ndarray
- Parameter that minimizes function.
- fopt : float
- Value of function at minimum:
fopt = func(xopt). - iter : int
- Number of iterations performed.
- funcalls : int
- Number of function calls made.
- warnflag : int
- 1 : Maximum number of function evaluations made. 2 : Maximum number of iterations reached.
- allvecs : list
- Solution at each iteration.
- minimize: Interface to minimization algorithms for multivariate
- functions. See the ‘Nelder-Mead’ method in particular.
Uses a Nelder-Mead simplex algorithm to find the minimum of function of one or more variables.
This algorithm has a long history of successful use in applications. But it will usually be slower than an algorithm that uses first or second derivative information. In practice it can have poor performance in high-dimensional problems and is not robust to minimizing complicated functions. Additionally, there currently is no complete theory describing when the algorithm will successfully converge to the minimum, or how fast it will if it does.
[1] Nelder, J.A. and Mead, R. (1965), “A simplex method for function minimization”, The Computer Journal, 7, pp. 308-313 [2] Wright, M.H. (1996), “Direct Search Methods: Once Scorned, Now Respectable”, in Numerical Analysis 1995, Proceedings of the 1995 Dundee Biennial Conference in Numerical Analysis, D.F. Griffiths and G.A. Watson (Eds.), Addison Wesley Longman, Harlow, UK, pp. 191-208.
fmin_powell¶
-
popeye.utilities.fmin_powell(func, x0, args=(), xtol=0.0001, ftol=0.0001, maxiter=None, maxfun=None, full_output=0, disp=1, retall=0, callback=None, direc=None)¶ Minimize a function using modified Powell’s method. This method only uses function values, not derivatives.
- func : callable f(x,*args)
- Objective function to be minimized.
- x0 : ndarray
- Initial guess.
- args : tuple, optional
- Extra arguments passed to func.
- callback : callable, optional
- An optional user-supplied function, called after each
iteration. Called as
callback(xk), wherexkis the current parameter vector. - direc : ndarray, optional
- Initial direction set.
- xtol : float, optional
- Line-search error tolerance.
- ftol : float, optional
- Relative error in
func(xopt)acceptable for convergence. - maxiter : int, optional
- Maximum number of iterations to perform.
- maxfun : int, optional
- Maximum number of function evaluations to make.
- full_output : bool, optional
- If True, fopt, xi, direc, iter, funcalls, and warnflag are returned.
- disp : bool, optional
- If True, print convergence messages.
- retall : bool, optional
- If True, return a list of the solution at each iteration.
- xopt : ndarray
- Parameter which minimizes func.
- fopt : number
- Value of function at minimum:
fopt = func(xopt). - direc : ndarray
- Current direction set.
- iter : int
- Number of iterations.
- funcalls : int
- Number of function calls made.
- warnflag : int
- Integer warning flag:
- 1 : Maximum number of function evaluations. 2 : Maximum number of iterations.
- allvecs : list
- List of solutions at each iteration.
- minimize: Interface to unconstrained minimization algorithms for
- multivariate functions. See the ‘Powell’ method in particular.
Uses a modification of Powell’s method to find the minimum of a function of N variables. Powell’s method is a conjugate direction method.
The algorithm has two loops. The outer loop merely iterates over the inner loop. The inner loop minimizes over each current direction in the direction set. At the end of the inner loop, if certain conditions are met, the direction that gave the largest decrease is dropped and replaced with the difference between the current estiamted x and the estimated x from the beginning of the inner-loop.
The technical conditions for replacing the direction of greatest increase amount to checking that
- No further gain can be made along the direction of greatest increase from that iteration.
- The direction of greatest increase accounted for a large sufficient fraction of the decrease in the function value from that iteration of the inner loop.
Powell M.J.D. (1964) An efficient method for finding the minimum of a function of several variables without calculating derivatives, Computer Journal, 7 (2):155-162.
Press W., Teukolsky S.A., Vetterling W.T., and Flannery B.P.: Numerical Recipes (any edition), Cambridge University Press
gradient_descent_search¶
-
popeye.utilities.gradient_descent_search(parameters, bounds, data, error_function, objective_function, verbose)¶ A generic gradient-descent error minimization function.
The values inside parameters are used as a seed-point for a gradient-descent error minimization procedure [1]_. The user must also supply an objective_function for producing a model prediction and an error_function for relating that prediction to the actual, measured data.
In addition, the user may also supply fit_bounds, containing pairs of upper and lower bounds for each of the values in parameters. If fit_bounds is specified, the error minimization procedure in error_function will return an Inf whether the parameters exceed the minimum or maxmimum values specified in fit_bounds.
- parameters : tuple
- A tuple of values representing a model setting.
- args : tuple
- Extra arguments to objective_function beyond those in parameters.
- fit_bounds : tuple
- A tuple containing the upper and lower bounds for each parameter in parameters. If a parameter is not bounded, simply use None. For example, fit_bounds=((0,None),(-10,10),) would bound the first parameter to be any positive number while the second parameter would be bounded between -10 and 10.
- data : ndarray
- The actual, measured time-series against which the model is fit.
- error_function : callable
- The error function that relates the model prediction to the measure data. The error function returns a float that represents the residual sum of squared errors between the prediction and the data.
- objective_function : callable
- The objective function that takes parameters and args and proceduces a model time-series.
- estimate : tuple
- The model solution given parameters and objective_function.
[1] Fletcher, R, Powell, MJD (1963) A rapidly convergent descent method for minimization, Compututation Journal 6, 163-168.
inv¶
-
popeye.utilities.inv(a, overwrite_a=False, check_finite=True)¶ Compute the inverse of a matrix.
- a : array_like
- Square matrix to be inverted.
- overwrite_a : bool, optional
- Discard data in a (may improve performance). Default is False.
- check_finite : boolean, optional
- Whether to check the input matrixes contain only finite numbers. Disabling may give a performance gain, but may result to problems (crashes, non-termination) if the inputs do contain infinities or NaNs.
- ainv : ndarray
- Inverse of the matrix a.
- LinAlgError :
- If a is singular.
- ValueError :
- If a is not square, or not 2-dimensional.
>>> a = np.array([[1., 2.], [3., 4.]]) >>> sp.linalg.inv(a) array([[-2. , 1. ], [ 1.5, -0.5]]) >>> np.dot(a, sp.linalg.inv(a)) array([[ 1., 0.], [ 0., 1.]])
isscalar¶
-
popeye.utilities.isscalar(num)¶ Returns True if the type of num is a scalar type.
- num : any
- Input argument, can be of any type and shape.
- val : bool
- True if num is a scalar type, False if it is not.
>>> np.isscalar(3.1) True >>> np.isscalar([3.1]) False >>> np.isscalar(False) True
multiprocess_bundle¶
-
popeye.utilities.multiprocess_bundle(Fit, model, data, grids, bounds, Ns, indices, auto_fit, verbose)¶
normalize¶
-
popeye.utilities.normalize(array, imin=-1, imax=1, axis=-1)¶ A short-hand function for normalizing an array to a desired range.
- array : ndarray
- An array to be normalized.
- imin : float
- The desired minimum value in the output array. Default: -1
- imax : float
- The desired maximum value in the output array. Default: 1
- array : ndarray
- The normalized array with imin and imax as the minimum and maximum values.
ones¶
-
popeye.utilities.ones(shape, dtype=None, order='C')¶ Return a new array of given shape and type, filled with ones.
- shape : int or sequence of ints
- Shape of the new array, e.g.,
(2, 3)or2. - dtype : data-type, optional
- The desired data-type for the array, e.g., numpy.int8. Default is numpy.float64.
- order : {‘C’, ‘F’}, optional
- Whether to store multidimensional data in C- or Fortran-contiguous (row- or column-wise) order in memory.
- out : ndarray
- Array of ones with the given shape, dtype, and order.
zeros, ones_like
>>> np.ones(5) array([ 1., 1., 1., 1., 1.])
>>> np.ones((5,), dtype=np.int) array([1, 1, 1, 1, 1])
>>> np.ones((2, 1)) array([[ 1.], [ 1.]])
>>> s = (2,2) >>> np.ones(s) array([[ 1., 1.], [ 1., 1.]])
parallel_fit¶
-
popeye.utilities.parallel_fit(args)¶ This is a convenience function for parallelizing the fitting procedure. Each call is handed a tuple or list containing all the necessary inputs for instantiaing a GaussianFit class object and estimating the model parameters.
- args : list/tuple
- A list or tuple containing all the necessary inputs for fitting the Gaussian pRF model.
- fit : Fit class object
- A fit object that contains all the inputs and outputs of the pRF model estimation for a single voxel.
peakdet¶
-
popeye.utilities.peakdet(v, delta, x=None)¶ Converted from MATLAB script at http://billauer.co.il/peakdet.html
Returns two arrays
function [maxtab, mintab]=peakdet(v, delta, x) #PEAKDET Detect peaks in a vector # [MAXTAB, MINTAB] = PEAKDET(V, DELTA) finds the local # maxima and minima (“peaks”) in the vector V. # MAXTAB and MINTAB consists of two columns. Column 1 # contains indices in V, and column 2 the found values. # # With [MAXTAB, MINTAB] = PEAKDET(V, DELTA, X) the indices # in MAXTAB and MINTAB are replaced with the corresponding # X-values. # # A point is considered a maximum peak if it has the maximal # value, and was preceded (to the left) by a value lower by # DELTA. # # Eli Billauer, 3.4.05 (Explicitly not copyrighted). # This function is released to the public domain; Any use is allowed.
percent_change¶
-
popeye.utilities.percent_change(ts, ax=-1)¶ Returns the % signal change of each point of the times series along a given axis of the array timeseries
- ts : ndarray
- an array of time series
- ax : int, optional (default to -1)
- the axis of time_series along which to compute means and stdevs
- ndarray
- the renormalized time series array (in units of %)
>>> np.set_printoptions(precision=4) # for doctesting >>> ts = np.arange(4*5).reshape(4,5) >>> ax = 0 >>> percent_change(ts,ax) array([[-100. , -88.2353, -78.9474, -71.4286, -65.2174], [ -33.3333, -29.4118, -26.3158, -23.8095, -21.7391], [ 33.3333, 29.4118, 26.3158, 23.8095, 21.7391], [ 100. , 88.2353, 78.9474, 71.4286, 65.2174]]) >>> ax = 1 >>> percent_change(ts,ax) array([[-100. , -50. , 0. , 50. , 100. ], [ -28.5714, -14.2857, 0. , 14.2857, 28.5714], [ -16.6667, -8.3333, 0. , 8.3333, 16.6667], [ -11.7647, -5.8824, 0. , 5.8824, 11.7647]])
randn¶
-
popeye.utilities.randn(d0, d1, ..., dn)¶ Return a sample (or samples) from the “standard normal” distribution.
If positive, int_like or int-convertible arguments are provided, randn generates an array of shape
(d0, d1, ..., dn), filled with random floats sampled from a univariate “normal” (Gaussian) distribution of mean 0 and variance 1 (if any of the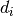 are
floats, they are first converted to integers by truncation). A single
float randomly sampled from the distribution is returned if no
argument is provided.
are
floats, they are first converted to integers by truncation). A single
float randomly sampled from the distribution is returned if no
argument is provided.This is a convenience function. If you want an interface that takes a tuple as the first argument, use numpy.random.standard_normal instead.
- d0, d1, ..., dn : int, optional
- The dimensions of the returned array, should be all positive. If no argument is given a single Python float is returned.
- Z : ndarray or float
- A
(d0, d1, ..., dn)-shaped array of floating-point samples from the standard normal distribution, or a single such float if no parameters were supplied.
random.standard_normal : Similar, but takes a tuple as its argument.
For random samples from
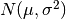 , use:
, use:sigma * np.random.randn(...) + mu>>> np.random.randn() 2.1923875335537315 #random
Two-by-four array of samples from N(3, 6.25):
>>> 2.5 * np.random.randn(2, 4) + 3 array([[-4.49401501, 4.00950034, -1.81814867, 7.29718677], #random [ 0.39924804, 4.68456316, 4.99394529, 4.84057254]]) #random
recast_estimation_results¶
-
popeye.utilities.recast_estimation_results(output, grid_parent, overloaded=False)¶
romb¶
-
popeye.utilities.romb(y, dx=1.0, axis=-1, show=False)¶ Romberg integration using samples of a function.
- y : array_like
- A vector of
2**k + 1equally-spaced samples of a function. - dx : array_like, optional
- The sample spacing. Default is 1.
- axis : array_like?, optional
- The axis along which to integrate. Default is -1 (last axis).
- show : bool, optional
- When y is a single 1-D array, then if this argument is True print the table showing Richardson extrapolation from the samples. Default is False.
- ret : array_like?
- The integrated result for each axis.
quad - adaptive quadrature using QUADPACK romberg - adaptive Romberg quadrature quadrature - adaptive Gaussian quadrature fixed_quad - fixed-order Gaussian quadrature dblquad, tplquad - double and triple integrals simps, trapz - integrators for sampled data cumtrapz - cumulative integration for sampled data ode, odeint - ODE integrators
seed¶
-
popeye.utilities.seed(seed=None)¶ Seed the generator.
This method is called when RandomState is initialized. It can be called again to re-seed the generator. For details, see RandomState.
- seed : int or array_like, optional
- Seed for RandomState. Must be convertible to 32 bit unsigned integers.
RandomState
shuffle¶
-
popeye.utilities.shuffle(self, x, random=None, int=<type 'int'>)¶ x, random=random.random -> shuffle list x in place; return None.
Optional arg random is a 0-argument function returning a random float in [0.0, 1.0); by default, the standard random.random.
solve¶
-
popeye.utilities.solve(a, b, sym_pos=False, lower=False, overwrite_a=False, overwrite_b=False, debug=False, check_finite=True)¶ Solve the equation
a x = bforx.- a : (M, M) array_like
- A square matrix.
- b : (M,) or (M, N) array_like
- Right-hand side matrix in
a x = b. - sym_pos : bool
- Assume a is symmetric and positive definite.
- lower : boolean
- Use only data contained in the lower triangle of a, if sym_pos is true. Default is to use upper triangle.
- overwrite_a : bool
- Allow overwriting data in a (may enhance performance). Default is False.
- overwrite_b : bool
- Allow overwriting data in b (may enhance performance). Default is False.
- check_finite : boolean, optional
- Whether to check the input matrixes contain only finite numbers. Disabling may give a performance gain, but may result to problems (crashes, non-termination) if the inputs do contain infinities or NaNs.
- x : (M,) or (M, N) ndarray
- Solution to the system
a x = b. Shape of the return matches the shape of b.
- LinAlgError
- If a is singular.
Given a and b, solve for x:
>>> a = np.array([[3,2,0],[1,-1,0],[0,5,1]]) >>> b = np.array([2,4,-1]) >>> x = linalg.solve(a,b) >>> x array([ 2., -2., 9.]) >>> np.dot(a, x) == b array([ True, True, True], dtype=bool)
spm_hrf¶
-
popeye.utilities.spm_hrf(delay, TR)¶ An implementation of spm_hrf.m from the SPM distribution
Arguments:
Required: TR: repetition time at which to generate the HRF (in seconds)
Optional: p: list with parameters of the two gamma functions:
defaults(seconds)
p[0] - delay of response (relative to onset) 6 p[1] - delay of undershoot (relative to onset) 16 p[2] - dispersion of response 1 p[3] - dispersion of undershoot 1 p[4] - ratio of response to undershoot 6 p[5] - onset (seconds) 0 p[6] - length of kernel (seconds) 32
trapz¶
-
popeye.utilities.trapz(y, x=None, dx=1.0, axis=-1)¶ Integrate along the given axis using the composite trapezoidal rule.
Integrate y (x) along given axis.
- y : array_like
- Input array to integrate.
- x : array_like, optional
- The sample points corresponding to the y values. If x is None, the sample points are assumed to be evenly spaced dx apart. The default is None.
- dx : scalar, optional
- The spacing between sample points when x is None. The default is 1.
- axis : int, optional
- The axis along which to integrate.
- trapz : float
- Definite integral as approximated by trapezoidal rule.
sum, cumsum
Image [2]_ illustrates trapezoidal rule – y-axis locations of points will be taken from y array, by default x-axis distances between points will be 1.0, alternatively they can be provided with x array or with dx scalar. Return value will be equal to combined area under the red lines.
[1] Wikipedia page: http://en.wikipedia.org/wiki/Trapezoidal_rule [2] Illustration image: http://en.wikipedia.org/wiki/File:Composite_trapezoidal_rule_illustration.png >>> np.trapz([1,2,3]) 4.0 >>> np.trapz([1,2,3], x=[4,6,8]) 8.0 >>> np.trapz([1,2,3], dx=2) 8.0 >>> a = np.arange(6).reshape(2, 3) >>> a array([[0, 1, 2], [3, 4, 5]]) >>> np.trapz(a, axis=0) array([ 1.5, 2.5, 3.5]) >>> np.trapz(a, axis=1) array([ 2., 8.])
zscore¶
-
popeye.utilities.zscore(time_series, axis=-1)¶ Returns the z-score of each point of the time series along a given axis of the array time_series.
- time_series : ndarray
- an array of time series
- axis : int, optional
- the axis of time_series along which to compute means and stdevs
- zt : ndarray
- the renormalized time series array